The Molecules Gateway lists 150,777 different entries. Of these, 1,031 are present in the seven different unfermented media used to cultivate the strains and derive from the complex ingredients (i.e. soy peptone, soluble starch, casein hydrolysate, yeast extract, meat extract, soybean meal and bacto-peptone ) used for media preparation. These molecules are labeled as such in the Molecules Gateway.
Annotation levels and annotation tools

The three annotation tools – Compound Discoverer (CD), MolDiscovery (MD) and MS2Query (MQ) – predicted molecules at very different rates, ranging from over one third of entries for CD to just 4% for MD. Of note, molecule prediction by a tool does not imply that the prediction is correct.

Frequency of molecules
Frequently occurring molecules are expected to represent medium components, molecules from primary metabolism or common specialized metabolites. Most molecules are present in a few extracts only, and only 3,120 molecules are contained in more than 200 extracts. See the frequency of molecules present in the 1–200 extract range.

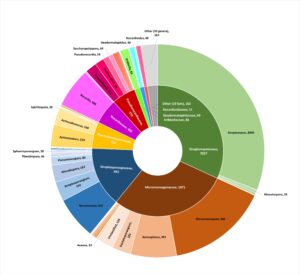
Taxonomic origin

Molecular diversity
How different are the molecules listed? This question can be answered, by looking at the chemical relatedness and originating biosynthetic pathway for the 5,660 unique InChIKeys listed in the Molecules Gateway (1417 molecules arranged into families and 4243 molecules forming single nodes), and at the distribution of exact mass and retention time for the 58,093 molecules with 1 through 7 annotation confidence level. These analyses indicate that all major biosynthetic pathways are represented, that a limited number of closely related molecular families occurs and that there is no obvious bias in retention time or molecular weight.


